Using the command line
Kernel Density Estimation
Most steps that are available from the GUI can also be called from the command line.
library(rhr)
data(datSH)
str(datSH)## 'data.frame': 1500 obs. of 5 variables:
## $ collar : chr "c5182" "c5182" "c5182" "c5182" ...
## $ x_epsg31467: int 3558403 3558548 3558541 3558453 3558566 3557836 3557881 3557150 3557021 3555975 ...
## $ y_epsg31467: int 5999400 5999099 5999019 5999026 5999365 5999185 5999139 5999159 5998865 5997615 ...
## $ day : chr "2009-02-13" "2009-02-13" "2009-02-13" "2009-02-13" ...
## $ time : chr "00:02:23" "06:02:21" "12:01:51" "18:00:55" ...We see that x and y coordinates are saved in the second and third column. Next we will perform a KDE.
kd1 <- rhrKDE(datSH[, 2:3])## rhrRasterFromExt: using equal nrow and ncolThe kde functions take three additional arguments: bandwidth, template raster and levels.
Bandwidth
The bandwidth can be provided as a vector of length one or two. Several methods to estimate bandwidth are available.
rhrHref: estimates the reference bandwidth.rhrHlscv: estimates the least square cross validation bandwidth.rhrHpi: estimates the plug in the equation bandwidth.rhrHrefScaled: estimates a scaled re fence bandwidth (as described in @Kie2013)
The default bandwidth is the reference bandwidth.
## Calculating reference bandwidth:
(href <- rhrHref(datSH[, 2:3]))## $h
## [1] 618.104 618.104
##
## $rescale
## [1] "none"## Or plugin the equation
(hpi <- rhrHpi(datSH[, 2:3]))## $h
## [1] 65.04826 74.39719
##
## $rescale
## [1] "none"
##
## $correct
## [1] TRUEkdehref1 <- rhrKDE(datSH[, 2:3], h = href$h)## rhrRasterFromExt: using equal nrow and ncolkdehref2 <- rhrKDE(datSH[, 2:3], h = rhrHref(datSH[, 2:3])$h)## rhrRasterFromExt: using equal nrow and ncol## Are they the same
identical(rhrUD(kdehref1), rhrUD(kdehref2))## [1] TRUEWorking with the result
Once we successfully estimated a KDE, there are several things we can do with. The first thing might be to plot it:
plot(kdehref1)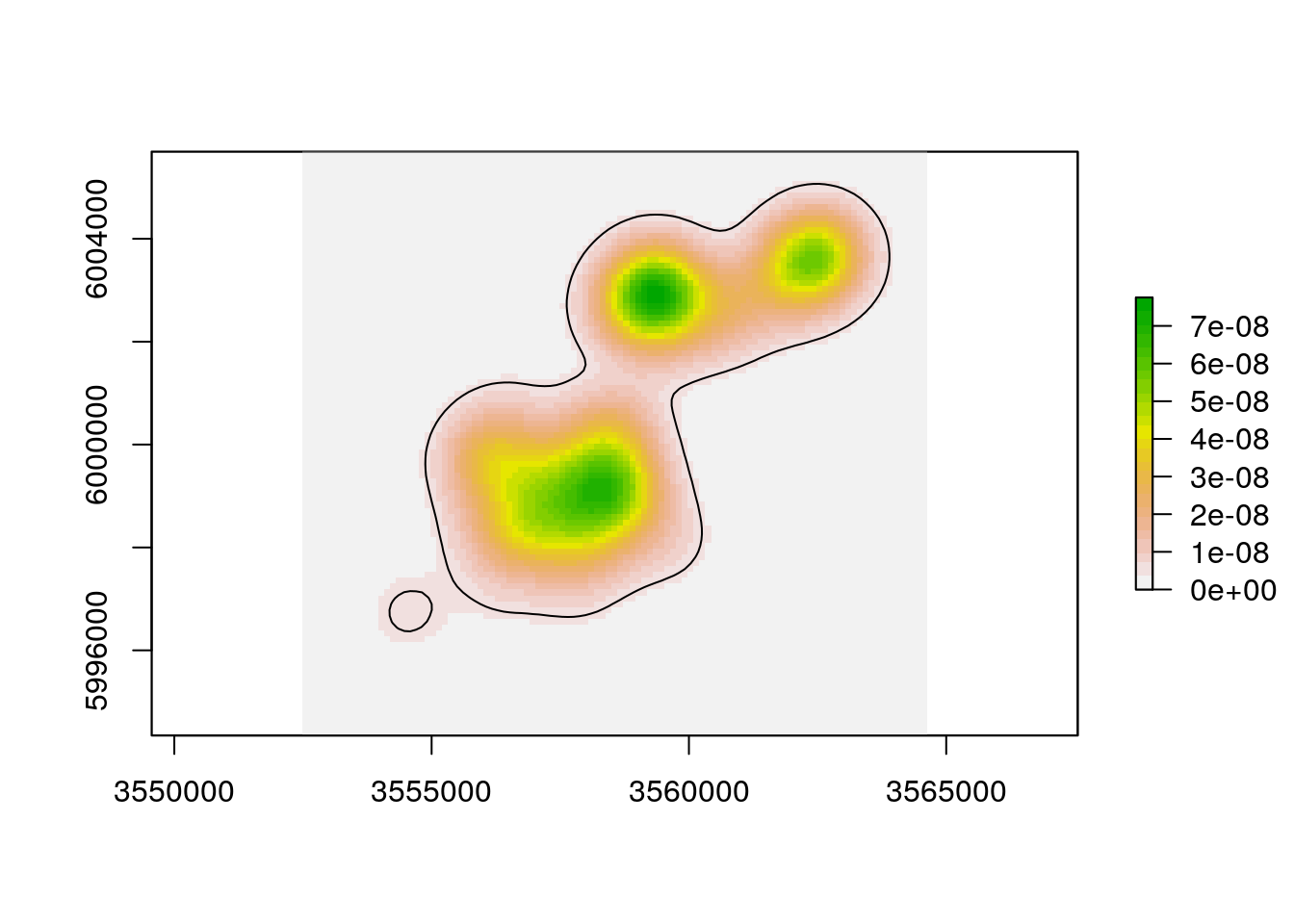
The functions rhrUD and rhrIsopleths allows to retrieve the UD and isopleths respectively.
ud <- rhrUD(kdehref1)
class(ud)## [1] "RasterLayer"
## attr(,"package")
## [1] "raster"The ud is a RasterLayer from the raster package. Similarly we can obtain the the isopleths at a predefined or specific level.
## the default level of 95
iso1 <- rhrIsopleths(kdehref1)
plot(iso1)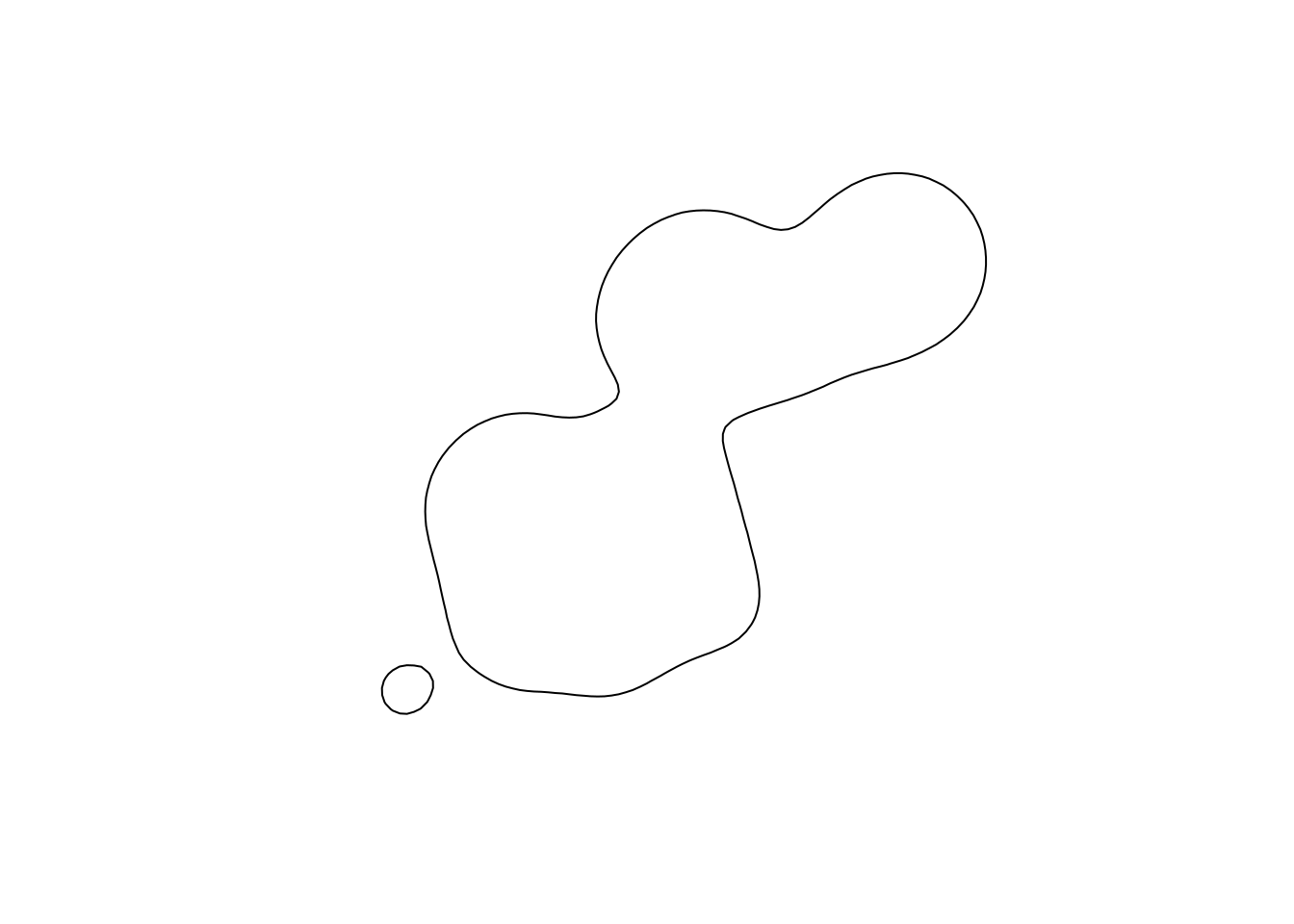
class(iso1)## [1] "SpatialPolygonsDataFrame"
## attr(,"package")
## [1] "sp"## other levels
iso2 <- rhrIsopleths(kdehref1, levels = seq(10, 90, 5))
plot(iso2) The
The rhrArea functions returns the home-range area at a given isopleth.
## default level
rhrArea(kdehref1)## level area
## 1 95 38325055## other levels
rhrArea(kdehref1, levels = seq(10, 90, 5))## level area
## 1 10 1401063
## 2 15 2203714
## 3 20 3089250
## 4 25 4019084
## 5 30 5011304
## 6 35 6069462
## 7 40 7215234
## 8 45 8435629
## 9 50 9771290
## 10 55 11211624
## 11 60 12819124
## 12 65 14571133
## 13 70 16664247
## 14 75 18931113
## 15 80 21714672
## 16 85 25249938
## 17 90 30171443Saving results
Isopleths can be saved various GIS formats, e.g.,
raster::shapefile(iso1, "myShape.shp")Working with several animals/instances
Often it may be useful to work with more than one animal or instance (i.e., different tracking periods of the same animal) simultaneously. rhr does not provide direct support for this. However, it can be easily achieved using base R.
As an example, we will estimate monthly home ranges for the example red deer data set datSH. To do this we have to go through 3 steps:
- Define a factor to split the data (e.g., species id, month of the year, study area).
- Split the data with R’s
splitfunction. - Apply some home range estimate.
library(rhr)
library(lubridate)
data(datSH)
# 1. define a splitting variable, here I will use the months
datSH$month <- round_date(ymd(datSH$day), "month")
# 2. Split the data
dat2 <- split(datSH, datSH$month)
# 3. Apply home range estimates
hrs <- lapply(dat2, function(x) rhrKDE(x[, 2:3])) # x is now the data for 1 month## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncolhrs is now a list where each entry is the kernel density estimate for one month. To obtain the home range size (HRS) of the first month, we can use:
rhrArea(hrs[[1]])## level area
## 1 95 774638and with sapply we can easily obtain the home range size for all months.
sapply(hrs, rhrArea)## 2008-04-01 2008-05-01 2008-06-01 2008-07-01 2008-08-01 2009-01-01
## level 95 95 95 95 95 95
## area 774638 9727988 21492189 5226063 3799606 502955.9
## 2009-02-01 2009-03-01 2009-04-01 2009-05-01
## level 95 95 95 95
## area 7797540 15470853 7508145 7501840Adjusting animal specific parameters
If, e.g., parameters such as the bandwidth should be instance specific there are two ways:
- Estimate the parameters for each instance.
- Estimate the parameter on the fly.
Adjusting the example from above we get:
library(rhr)
library(lubridate)
data(datSH)
dat2 <- split(datSH, round_date(ymd(datSH$day), "month"))
bw <- lapply(dat2, function(x) rhrHpi(x[, 2:3]))
hrs <- lapply(seq_along(bw), function(i) rhrKDE(dat2[[i]][, 2:3], h = bw[[i]]$h)) ## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncolWe could also do this on the fly with
dat2 <- split(datSH, round_date(ymd(datSH$day), "month"))
hrs <- lapply(dat2, function(x) rhrKDE(x[, 2:3], h = rhrHpi(x[, 2:3])$h)) ## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol
## rhrRasterFromExt: using equal nrow and ncol